The 2014 Ebola outbreak in West Africa captured the nation’s attention, highlighting the need for improved biosurveillance at the border and identifying a capability gap in the United States’ homeland defense and security architecture. In response, the U.S. strove to fill knowledge gaps and operational capabilities by advancing experimental patient treatments and therapies, strengthening the traveler screening process, and enabling a national strategic response framework for infectious disease— working with the United Nations and states of Liberia, Sierra Leone, and Guinea to contain the epidemic [1]. Through the execution of Operation United Assistance, the U.S. Department of Defense (DoD) dedicated more than $2.3 billon to the deployment of medical military personnel, research labs, hospital beds, and other necessary medical supplies to help more than 28,000 West Africans affected by the outbreak [2]. In addition, the Centers for Disease Control and Prevention (CDC), Department of Homeland Security (DHS), and West African governments implemented traveler health screenings at ports of entry (POE) to fortify border security and contain the spread of the virus.
To enact a risk-based screening process at domestic POEs (see Figure 1), officers from U.S. Customs and Border Protection (CBP) assessed travelers entering the country for potential exposure to Ebola. High-risk travelers—including those entering from the three affected African states [3]—received an enhanced health screening, in which a non-contact infrared thermometer was used to measure their temperature. Travelers with temperatures greater than 100 degrees Fahrenheit were assessed further and then advised to seek medical care [4–10].
In October 2014, after CDC confirmed the first case of Ebola in the United States, CBP began to implement an enhanced entry screening program. In the first 30 days of the program, CBP officers screened 1,993 travelers from Guinea, Liberia, and Sierra Leone. CBP referred 86 of those travelers to the CDC for a more intensive health screening [3]. Of those, seven asymptomatic individuals were sent for further medical evaluation [3]. While deployed, U.S. military personnel serving in Operation United Assistance were also monitored daily for exposure symptoms [11, 12].
Before re-entering the U.S., non-symptomatic DoD personnel received additional medical screening and were placed in controlled monitoring for 21 days [11, 12]. During this time, military personnel with no known exposure were assessed for febrile symptoms, measuring their temperature twice daily [12]. DoD civilians supporting the deployments (but without known exposure) were permitted to either voluntarily participate in DoD’s controlled monitoring protocol, or follow guidance from CDC, state, and local public health authorities on the active monitoring of Ebola [12].
During the outbreak, border screenings of travelers were essential to preventing the biothreat from spreading to other countries [13]. More than 38,000 travelers entering the U.S. underwent enhanced Ebola screening [14]. Apart from the results of traveler questionnaires taken during CBP’s secondary screening stage, the only other determining risk factors were symptoms of infection—namely an elevated temperature. Of the 38,000 travelers screened in this manner, 2,975 returned positive responses to the questionnaire, 81 travelers presented elevated temperatures, 2,996 travelers were referred to tertiary screening, and 49 travelers were sent to medical facilities for further assessment [4].
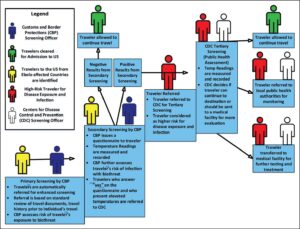
Figure 1. Risk-based screening process for travelers entering the U.S. During the 2014 Ebola Outbreak. This figure demonstrates the traveler screening process used to assess travelers to the United States during the 2014 Ebola outbreak [4–10].
However, the only diagnostic tool currently available for secondary screening in CBP’s tiered process is a thermometer. The Ebola outbreak reinforced the need to deliver a field-deployable diagnostic test capable of rapidly and non-invasively screening pre-symptomatic travelers for diseases of concern.
Development of a diagnostic tool for this stage could expedite the screening process, improve response times for positive tests, and provide public health officials with better information than current questionnaire- and temperature-based tools. Moreover, with a severe biothreat like Ebola, the use of non-invasive diagnostics is critical for protecting medical screening officers (and other travelers) from exposure. Non-invasive sampling collection techniques are defined as such because they do not penetrate through the skin. Examples include nasal, oral, and epidermal swabs, or the collection of breath samples.
Required characteristics for an effective POE screening device include: a high degree of portability, user friendliness, durability, non-invasive sample collection, generation of minimal hazardous material during the analysis process, accurate diagnostics for multiple diseases in less than 60 seconds (a standard benchmark for “rapid” analysis), and the ability to detect a pathogen in an asymptomatic traveler.
This article reviews four distinct technologies that can be applied to improving research and development (R&D) in the field of rapid diagnostics. These four technologies include nucleic acid-based detection technology; antigen-antibody binding-based detection technology; volatile organic compound-based detection technology; and infrared light-based detection technology. A review of the literature suggests that current R&D for improving the performance of rapid diagnostic tests is focused heavily on tests rooted in nucleic acid-based and antigen-antibody binding-based detection technologies (see Figure 2) [16–19]. Less emphasis and attention have been paid to the development of volatile organic compound-based and infrared light-based detection technologies.
Testing for Diseases of Public Health Significance
Four classes of disease can be closely monitored and detected using traditional screening practices (e.g., measuring temperature, heart rate, or other symptoms). These four classes include infectious respiratory, viral hemorrhagic, vector-borne, and gastrointestinal disease. Foreign nationals seeking to enter the U.S. under immigration or refugee protocols must pass a medical examination as part of the visa application process, and those who present symptoms of what CDC terms “communicable diseases of public health significance” are excludable from entry [20]. Foreign nationals visiting or transiting through the U.S. who present symptoms of such diseases may also be examined before entry is granted. U.S. citizens re-entering the country are not required to pass a health screening to gain entry, except in the event of an emerging disease outbreak posing a significant threat to the nation’s public health.
Diagnostic tests vary in their approach to disease detection, but many are designed to target an analyte specific to a pathogen that can be bound to some form of a bioreceptor. After binding, a transducer can be used to amplify a signal that indicates the binding of the analyte to the receptor. Diagnostic tests designed in this manner are crucial for pathogen detection in vitro and allow medical professionals to select an appropriate treatment regimen. Diagnostic technologies also vary in their processing times, instrumentation reliability, and tradeoffs between specificity and sensitivity.
Nucleic Acid-based Detection
Nucleic acid-based detection technologies use genetic code to identify pathogens. While doing so, nucleic acid-based detectors will target chemical compounds within either deoxyribonucleic acid (DNA) or ribonucleic acid (RNA). Several technologies, such as reverse transcriptase-polymerase chain reaction (RT-PCR), PCR, and real-time PCR (qPCR), can detect and confirm multiple pathogens through nucleic acid amplification and quantification. Many Ebola diagnostic assays used during the 2014 outbreak were developed on the principles of RT-PCR [21]. While these lab-based methods are the most accurate available, they require expensive instrumentation, a high degree of technical expertise, invasive sampling techniques (e.g., venipuncture), and lengthy processing times (two or more hours) to operate. However, nucleic acid amplification provides a thorough analysis of the amplified products. Targeted amplification, combined with nucleic acid sequencing, can provide more detail about a biological agent, potentially including its relationship to another biothreat, or its susceptibility to medical countermeasures.
Through technical R&D, the diagnostics industry has improved our ability to complete sequencing in a field-deployable (non-laboratory) environment. For example, Oxford Nanopore Technologies Ltd. developed the MinION Nanopore Sequencer, which shows promise for use at POE. The MinION has demonstrated its ability to rapidly sequence viral genomes during the early phases of an influenza pandemic [22] and during the Zika virus epidemic [23]; and it was used by researchers in Guinea during the Ebola outbreak to process serum samples from patients in real time [24]. Powered by a USB port, the MinION relies on a flow cell to analyze nucleotide bases and it can simultaneously run multiple samples. The sequencer can operate independent of an internet connection and process a sample within an hour. Using the MinION, researchers have conducted real-time bioinformatics analysis in resource-limited areas such as the rain forest [25]. It has also been used in the rapid metagenomic detection of Chikungunya virus, Ebola, and hepatitis C virus in human blood samples [26]. These features make the MinION a promising candidate for border medical screening, if the requirement for non-invasive sample collection is waived.
Reverse transcriptase-recombinase polymerase amplification (RT-RPA) is another nucleic acid-based approach for pathogen detection. These assays function by placing enzymes and the DNA or RNA sample of interest within a centrifuge to initiate the amplification reaction (which can take place at room temperature). RT-RPAs have been useful in the on-site detection of dengue virus in human serum samples [27]; in the laboratory-based detection of multiple strains of Rift Valley fever virus taken from virus cultures [28]; and in the on-site detection of yellow fever virus taken from cultures, mosquito pools, and human plasma samples [29]. These assays benefit from the fact that the reaction does not require a thermocycler, which minimizes assay times and expense. These assays have also been used to identify viruses in pigs such as type 2 porcine reproductive and respirator syndrome virus [30], and detect epidemic human norovirus strains in viral RNA [31]. Reverse transcription loop-mediated isothermal amplification assays employ a similar technique, and have been proven to identify Middle East Respiratory Syndrome Coronavirus in human samples [32].
Synthetic biology—the generation of new biological products like genes, cells, or enzymes using advanced bioengineering techniques—has also furthered the development of nucleic acid-based detection technologies. For example, the development of programmable RNA sensors has led to the production of paper-based methods for virus detection. Such methods combine RNA sensors, known as Toehold Switches, with a freeze-dried, cell-free protein expression paper platform [33]. These methods are capable of identifying Zika virus without reporting a false detection of the closely related dengue virus [33]. As a review of the literature indicates, R&D on nucleic acid-based detection technology continues to be the primary focus in the advancement of rapid diagnostic tests.
Antigen-antibody Binding-based Detection
Relied on as diagnostic instruments since the early 1990s, antigen-antibody binding- based detection technologies work rapidly and are easy to interpret. The most relied-upon assays are Lateral Flow Assays (LFAs), which are relatively inexpensive and user-friendly [34]. Antigen-antibody binding diagnostic tests use blood proteins (antibodies) to capture foreign substances from pathogens (antigens). Antigen-antibody binding is specific; this means that only certain antibodies can bind to certain antigens. For a disease-detecting assay to function, strips are coated with pathogen-specific antibodies. Once a sample is applied, antigens flow toward the antibodies, which are coated in gold or latex particles. They bind and form complexes that accumulate in a line, yielding a colorimetric result that indicates the absence or presence of the pathogen. LFAs’ user-friendliness and cost-efficiency make them appealing tools for medical screening in non-laboratory environments.
However, LFAs can be limited in their analytical capabilities due to sensitivity and specificity challenges. Many LFA test strips are singleplex, or single-pathogen-specific. This is problematic when attempting to detect the presence of unidentified (or multiple) pathogens. Recent efforts have produced multiplex LFAs capable of detecting multiple pathogens simultaneously. Researchers have demonstrated the multiplex detection of 10 epidemic foodborne pathogens, including Escherichia coli, Salmonella, and Cholera [34]. However, multiplex LFAs face challenges like high levels of cross-reactivity (non-specific binding to antibodies), which can produce false positives among multiple pathogens [35]. Often used in environmental sampling, LFAs have identified the presence of biothreat agents like Bacillus anthracis [36]. Furthermore, these types of tests have been integrated with the use of mobile devices (e.g., smartphones, tablets) in analyzing results. For example, Biothreat Alert Multiplex Strips, produced by Tetracore, target up to five different antigen targets at once: a control line, Yersinia pestis, Francisella tularenis, Bacillus anthracis, and Burkholderia. These strips can be read and analyzed by Tetracore’s BTA Reader CX and BTA Reader TX phone or tablet add-ons to confirm the identity of the pathogens within 60 seconds [37].
Given their limitations regarding sample collection and multiplex-use, LFAs are only moderately suitable for the biomedical screening of travelers at POEs. In comparison to nucleic acid-based diagnostic assays, LFAs are more cost-effective and require less scientific expertise for use. However, when used outside of a laboratory environment, antigen-antibody tests are susceptible to factors that can compromise their reliability. Variables like temperature, light, and pH can compromise an LFA’s results. Assay performance is also affected by the shelf life and storage conditions of the testing reagents.
Volatile Organic Compound-based Detection
Defined in a medical context, volatile organic compounds (VOCs) are small molecules produced by the body (and/or microbial pathogens) and liberated in bodily fluids like breath, urine, feces, and sweat [38]. Research on the viability of using VOCs as indicators for infections is underway, but the scientific literature has already recognized an association of several VOC arrays with specific diseases. Urinary tract, gastro-intestinal, fungal, and bacterial infections, such as Mycobacterium tuberculosis, are just a few examples of instances where VOCs can be used as markers for the detection of disease [39]. Studies have also linked certain concentrations of acetone in breath to diabetes mellitus [40]; increased levels of hydrogen cyanide in the breath of patients aged seven to 17 to lung infections induced by Pseudomonas aerugnosa [41]; and the presence of thioethers in breath to Plasmodium falciparum, the protozoan parasite that causes malaria in humans [42].
In addition, several VOC-releasing bacterial pathogens—such as Staphylococcus aureus, Streptococcus pneumoniae, Enterobacter faecalis, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli—are being studied in order to produce statistical models of VOC– pathogen production relationships [43]. VOC analysis relies on the use of traditional chemical analytical technologies such as gas chromatography coupled with mass spectrometry (GC-MS) for compound identification. GC-MS is the gold standard for gas sample analysis, and it can analyze breath-borne compounds. Studies have used GC-MS in the detection of chemicals like ammonia, which, when present in breath, correlates with Helicobacter pylori infection in humans [44]. Other types of spectrometry that can be used to analyze chemical compounds include ion mobility spectrometry (IMS), selected ion flow tube mass spectrometry (SIFT-MS), and proton transfer spectrometry (PTS). IMS and SIFT-MS can provide real-time measurements of VOCs, and PTS has been used to detect VOCs associated with food spoilage-related bacteria [45]. All of these technologies provide a strong platform for the chemical analysis of gas samples.
Electronic noses, or e-noses, are sensors used to continually detect the presence of VOCs in gas samples [46]. E-nose technology relies on electronic sensors to identify patterns and recognize odors, but also typically integrates traditional chemical analyzers (like gas chromatography) into the system. Several e-nose devices are being improved upon or optimized for detecting the presence of disease-associated VOCs in human breath profiles [47]. Airborne chemical detection has already been deployed in efforts to interdict illegal substances and contraband at national
borders [48], and tools for VOC detection are showing promise for use in the interdiction of human trafficking activity [49]. Recognizing that canines’ exceptional sense of smell grants them the ability to sniff out certain chemicals and biomarkers, researchers have undertaken studies to better understand the flow of air within a dog’s nose, to inform the development of better e-nose devices [50, 51].
Analysis of VOCs in human breath shows significant potential for swiftly and accurately identifying travelers infected with a disease of concern during a medical screening. The release of VOCs in the gaseous phase makes these compounds a non-invasive means of sample collection [52, 53]. However, the release of relevant biomarker VOCs may not occur until a person is in the very late stages of infection, making detection challenging during the pre-symptomatic stage. As additional R&D focuses on exploring VOCs as markers for infectious disease, development of a portable, non-invasive VOC diagnostic tool may be plausible in the near future.
Infrared Light-based Detection
Like VOC sampling, infrared light also holds promise for use as a diagnostic technique—and it may be the most ideal technology for the development of a rapid, non-invasive detection device. Infrared light is often used in research laboratories as an analytical tool for determining the chemical components of solid or liquid samples. In comparison to ultraviolet radiation or visual light, infrared light reflects lower energy levels when applied to a sample [54], thus decreasing the risk of damage.
At present, no diagnostic test for an infectious disease uses this technology in vivo. However, infrared light-based detection devices have been deployed as biosurveillance scanners and used to detect febrile symptoms when screening travelers during outbreaks. Infrared thermal imaging scanners were used, with varying levels of effectiveness, to identify travelers presenting fevers during several seasonal flu outbreaks [55]; to complement other detection measures during the 2003 Severe Acute Respiratory Syndrome (SARS) outbreak [56]; and during the 2014 Ebola outbreak. The use of infrared thermal imaging scanners and non-contact infrared thermometers allows border officials to assess whether a given traveler may pose a risk of pathogen infection. However, it is important to note that non-contact thermometers have displayed limited efficacy in detecting the early stages of viral infections (such as influenza) in travelers [57].
Currently, engineers are working to apply this non-invasive technology to the development of user-friendly medical diagnostic instruments. In an ideal rapid and non-invasive diagnostic device, the instrument should be capable of directing light waves through the epidermal layer of the skin to detect the presence of a pathogen—without damaging the body. However, this diagnostic capability is not yet feasible, and the body of scientific literature related to its maturation remains limited in size. Direct application of infrared light to the body faces significant technical challenges, such as interference by dense physical features of the skin that inhibit optical analysis. Skin pigmentation and thickness all influence the propagation of light waves, which in turn impacts the accuracy of optical measurements [58].
While there is no light-based detection technology available for diagnosis in vivo, there is a laboratory technique used to analyze samples for bacterial and viral presence. This technique, known as Fourier transform infrared spectroscopy (FTIR), uses vibrational spectroscopy to examine vibrations between molecules. FTIR relies on infrared radiation to analyze the chemical composition of samples, and it is the vibrational spectroscopic technique most widely used in bacterial detection [59]. FTIR technology can identify structural information, like molecular changes to cellular components, allowing for the rapid identification of bacteria. To date, this technology has successfully detected E. coli O157:H7 in apple juice [60]; has been applied as a diagnostic and surveillance tool for cancer [61]; and has been demonstrated as a rapid technique for the identification of bacteria like P. aeruginosa in sputum samples [62]. In addition, FTIR has also successfully sensed foodborne pathogens such as Listeria monocytogenes [63].
Studies are still needed regarding the use of FTIR in rapid diagnostics in the biomedical field. Some researchers have had success using near-infrared light waves to monitor glucose levels in individuals with diabetes (or susceptible to it) [64]. Scientists have also attempted to use near-infrared light spectroscopy to detect and study viral or bacterial particles in model systems. In May 2018, scientists reported the use of near-infrared spectroscopy to detect Zika virus in Aedes aegypti mosquitoes [65]. Also, influenza and Ebola have been detected in serum samples via infrared light technology [66].
Because they are subject to the same challenges observed with nucleic acid-based, antigen-antibody binding-based, and VOCbased detection technologies, infrared light-based detection is still costly and not well-suited for field environments. Even so, infrared methods require no sample preparation, and results can be produced quickly in the form of spectral images. More research on integrating spectroscopy techniques with disease diagnosis procedures may lead to hybridized diagnostic devices capable of non-invasively testing for a disease-causing pathogen, and providing an accurate diagnosis for further action.
Conclusion
The greatest challenge in developing a rapid diagnostic tool is designing a non-invasive sampling technique that can reliably identify pathogen carriers before they show symptoms. Traditional oral or nasopharyngeal swab specimen collection remains the least invasive method currently available for sampling. However, this technique still subjects the sample to lengthy processing times. The use of swabs generates hazardous waste, further increasing the risk of exposure and the spread of disease. The operational environment at a POE requires safer techniques, less costly, more specific, and more portable technologies to improve the medical screening process of travelers deemed high-risk for exposure during a biological outbreak. The use of VOCs and infrared light-based detection devices hold the most promise for meeting these criteria.
Travelers entering the country and warfighters returning to the U.S. after serving in areas with ongoing infectious disease outbreaks require extensive health screenings to ensure their safety and to protect the country against an outbreak. Continued R&D in improved rapid and non-invasive diagnostic technologies may enhance our nation’s ability to prevent dangerous biothreats from entering the U.S., while providing timely and effective treatment for military and government personnel at risk of exposure.
Disclaimer
The research discussed in this article was supported in part by an appointment to the U.S. Department of Homeland Security (DHS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and DHS. ORISE is managed by ORAU for the DOE under contract number DESC0014664. All opinions expressed in this paper are the author’s and do not necessarily reflect the policies and views of DHS, DOE, or ORISE.
References
1. Fact Sheet: U.S. Response to the Ebola Epidemic in West Africa. The White House. Office of the Press Secretary. Retrieved from https://obamawhitehouse.archives.gov/thepress office/2014/09/16/fact-sheet-us-response-ebola-epidemic-west-africa
2. Joint and Coalition Operational Analysis.(2016). Operation UNITED ASSISTANCE: The DOD Response to Ebola in West Africa. Retrieve from http://www.jcs.mil/Portals/36/ Documents/Doctrine/ebola/OUA_report_jan2016.pdf
3. Brown, C.M., Aranas, A.E., Benenson, G.A., Brunette, G., Cetron, M., Chen, TH., Cohen, N.J… & Pesik, N. (2014). Airport Exit and Entry Screening for Ebola — August– November 10, 2014. Morbidity and Mortality Weekly Report, 63(49), 1163-1167. Retrieved from http://europepmc.org/backend/ptpmcrender.fcgi?accid=PMC4584540&blobtype=pdf
4. The Department of Homeland Security. (2016, June). Ebola Response. Retrieved from https://www.dhs.gov/archive/ebola-response
5. The Department of Homeland Security. (2005). Memorandum of Understanding between The Department of Health and Human Services and The Department of Homeland Security. Retrieved from https:// www.dco.uscg.mil/Portals/9/DCO%20Documents/5p/CSNCOE/MOU%20between%20the%20DHHS%20and%20DHS.pdf?ver=2017-08-08-081620-793
6. Office of Inspector General. (2016, January). DHS’ Ebola Response Needs Better Coordination, Training, and Execution. Retrieved from
7. Centers for Disease Control and Prevention. (2014, October). Enhanced Ebola Screening to Start at Five U.S. Airports and New Tracking Program for all People Entering U.S. from Ebola-affected Countries. Retrieved from https://www.cdc.gov/media/releases/2014/p1008-ebola-screening.html
8. Cohen, N.J., Brown, C.M., Alvarado-Ramy, F., Bair-Brake, H., Benenson, G.A., Chen, T.H., … & Cetron, M.S. (2016). Supplement: Travel and Border Health Measures to Prevent the International Spread of Ebola. MMWR, 65(3), 57-67. doi: 10.15585/mmwr.su6503a9
9. Virginia Department of Health. (2018). Ebola-Basic Airport Screening and Active Monitoring Protocal. Retrieved from http://www.vdh.virginia.gov/epidemiology/epidemiology-fact-sheets/
ebola-frequently-asked-questions/ebola-basic-airport-screening-and-active-monitoring-protocol/
10. DeVries, A., Talley, P., Sweet, K., Kline, S., Stinchfield, P., Tosh, P., & Danila, R. (2016). Development and Implementation of the Ebola Traveler Monitoring Program and Clinical Outcomes of Monitored Travelers during October-May 2015, Minnesota. PloS One, 11(12), 1-11. doi:10.1371/journal.pone.0166797
11. Cardile, A.P., Murray, C.K., Littell, C.T., Shah, N.J., Fandre, M.N., Drinkwater, D.C., … & Vento, T.J. (2015). Monitoring Exposure to Ebola and Health of U.S. Military Personnel Deployed in Support of Ebola Control Efforts-Liberia, October 25, 2014-February 27, 2015. MMWR, 64(25), 690-694.
12. The Department of Defense. Under Secretary of Defense for Personnel and Readiness, Wright, J. (2014). Memorandum for Under Secretaries of Defense. Retrieved from http://archive.defense.gov/home/features/2014/1014_ebola/docs/Pre-Post-Deployment-Training-Screening-Monitoring-Guidance-for-DoD.pdf
13. Centers for Disease Control and Prevention. Ebola (Ebola Virus Disease) 2014-2016 Ebola Outbreak in West Africa.Retrieved from www.cdc.gov/vhf/ebola/history/2014-2016-outbreak/index.html
14. Centers for Disease Control and Prevention. Ebola (Ebola Virus Disease) Cost of the Ebola Epidemic. Retrieved from www.cdc.gov/vhf/ebola/history/2014-2016-outbreak/
cost-of-ebola.html
15. Centers for Disease Control and Prevention. Ebola (Ebola Virus Disease) Transmission. Retrieved from https://www.cdc.gov/vhf/ebola/transmission/index.html
16. The University of Arizona. (2000). Antibody Structure. The Biology Project. Retrieved from http://www.biology.arizona.edu/immunology/tutorials/antibody/structure.html
17. National Center for Biotechnology Information. PubChem Compound Database;CID=768, https://pubchem.ncbi.nlm.nih.gov/compound/768
18. National Center for Biotechnology Information. PubChem Compound Database;CID=180, https://pubchem.ncbi.nlm.nih.gov/compound/180
19. The University of The State of New York. The State Education Department. (2006).Reference Tables for Physical Setting/Physics. Retrieved from http://www.p12.nysed.gov/assessment/reftable/physics-rt/physics06tbl.pdf
20. Centers for Disease Control and Prevention. (2017, October). Technical instructions for panel physicians and civil surgeons, introduction and background. Retrieved from
https://www.cdc.gov/immigrantrefugeehealth/exams/ti/panel/technical-instructions/panel-physicians/introduction-background.html
21. Cherpillod, P., Shibler, M., Vieille, G., Cordey S., Vetter, P., & Kaiser, L. (2016). Ebola virus disease diagnosis by real-time RTPCR: A comparative study of 11 different procedures. Journal of Clinical Virology, 77, 9-14. doi:10.1016/j.jcv.2016.01.017
22. Wang, J., Moore, N.E., Deng, Y.M., Eccles, D.A., & Hall, R.J. (2015). MinION Nanopore Sequencing of an Influenza Genome. Frontiers in Microbiology, 6, doi:10.3389/
fmicb.2015.00766
23. Quick, J., Grubaugh, N.D., Pullan, S.T., Claro, I.M., Smith, A.D., Gangvarapu, K., Oliveira, G., Loman, N.J. (2017). Multiplex PCR method for MinION and Illumina sequencing
of Zika and other virus genomes directly from clinical samples. Nature Protocols, 12(6), 1261-1276. doi: 10.1038/nprot.2017.066
24. Quick, J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L,… & Carroll, M.W. (2016). Real-time, portable genome sequencing for Ebola surveillance. Nature, 30(7589), 228–232. doi:10.1038/nature16996
25. Pomerantz, A., Peñafiel, N., Arteaga, A., Bustamante, L., Pichardo, F., Coloma, L.A.,… & Prost, S. (2017). Real-Time DNA barcoding in remote rainforest using Nanopore sequencing. bioRxiv 189159.doi:10.1101/189159
26. Greninger, A, Naccache, S, Federman, S, Yu, G, Mbala, P, Bres, C, … & Chiu, C. (2015). Rapid Metagenomic identification of viral pathogens in clinicalsamples by real-time nanopore sequencing analysis. Genome Medicine, 7(9), 9. doi:10.1186/s13073-015-0220-9
27. Abd, E.l .,Wahed, A., Patel, P., Faye, O., Thaloengsok ,S., Heidenreich, D., …& Weidmann, M. (2015). Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection. PLoS ONE, 10(6), e0129682. doi:10.1371/journal.pone.0129682
28. Euler, M., Wang, Y., Netnwich, O., Piepenburg, O, Hufert, F.T., & Weidmann, M.(2012). Recombinase polymerase amplification assay for rapid detection of Rift Valley fever virus. Journal of Clinical Virology, 54(4), 308–312. doi:10.1016/j.jcv.2012.05.006
29. Escadafal, C., Faye, O., Sall, A.A., Faye, O., Weidmann, M., Strohmeier, O., … & Patel, P. (2014) Rapid Molecular Assays for the Detection of Yellow Fever Virus in Low-Resource Settings. PLoS Negl Trop Dis, 8(3), e2730. doi:10.1371/journal.pntd.0002730
30. Wang, J., Yuan, W., Han, Q., Wang, J., Liu, L..(2017). Reverse transcription recombinase polymerase amplification assay for rapid detection of type 2 porcine reproductive and respiratory syndrome virus. Journal of Virological Methods, 243, 55-60. doi: /10.1016/j.jviromet.2017.01.017
31. Moore, M.D., and Jaykus, L.A. (2017). Development of a Recombinase Polymerase Amplification Assay for Detection of Epidemic Noroviruses. Scientific Reports, 7, 40244.doi:/10.1038/srep40244
32. Bhardra, S., Jiang, Y.S., Kumar, M.R., Johnson, R.F., Hensley, L.E., Ellington, A.D. (2015) Real-Time Sequence-Validated Loop-Mediated Isothermal Amplification Assays for Detection of Middle East Respiratory Syndrome Coronavirus (MERS-CoV). PLoS ONE, 10(4), e0123126. doi:10.1371/ journal
33. Pardee, K., Green, A.A., Takahashi, M.K., Braff, D., Lamber, G., Lee, J.W., … & Collins, J.J. (2016). Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular
Components. Cell, 165(5), 1255- 1266. doi:/10.1016/j.cell.2016.04.059
34. Zhao, Y., Wang, H., Zhang, P., Sun, C., Wang, X., Wang, X., … & Zhou L. (2016).Rapid multiplex detection of 10 foodborne pathogens with an upconverting phosphor technology based 10-channel lateral flow assay. Scientific Reports, 6, 21342.doi:10.1038/srep21342
35. Hanafiah, K.M., Arifin, N., Bustami, Y., Noordin, R., Garcia, M., & Anderson, D. (2017). Development of Multiplexed Infectious Diseases Lateral Flow Assays: Challenges and Oppertunities. Diagnostics, 7(51), 1-9.doi:10.3390/diagnostics7030051.
36. Ramage, J.G., Prentice, K.W., DePalma,P.L., Venkateswaran, K.S., Chivukula, S., Chapman, C., … & Pillai, S.P. (2016). Comprehensive Laboratory Evaluation of a Highly Specific Lateral Flow Assay for the Presumptive Identification of Bacillus Anthracis spores in Suspicious White Powders and Environmental Samples. Health Security, 14(5), 351-365. doi:10.1089/hs.2016.0041
37. O’Brien, T. (2018). BioThreat Alert® Reader. Retrieved from http://www.tetracore.com/bio-warfare/
38. Filipiak, W., Mochalski, P., Filipiak, A., Ager, C., Cumeras, R., Davis, C.E., … & Troppmair, J. (2016). A Compendium of Volatile Organic Compounds (VOCs) Released by Human Cell Lines. Current Medicinal Chemistry,23, 2112-2131. doi:10.2174/092986732366616031612505
39. Sethi, S., Nanda, R., & Chakraborty, T. (2013). Clinical Application of Volatile Organic Compound Analysis for Detecting Infectious Diseases. Clinical Microbiology Reviews, 26(3), 462-475. doi:10.1128/CMR.00020-13.
40. Saasa, V., Malwela, T., Beukes, M., Mokgotho, M., Liu, C.P., & Mwakikunga, B. (2018). Sensing Technologies for Detection of Acetone in Human Breath for Diabetes Diagnosis and Monitoring. Diagnostics, 8(1), 12. doi:10.3390/diagnostics8010012
41. Enderby, B. Smith, D. Carroll, W., & Lenney, W. (2009). Hydrogen cyanide as a biomarker for Pseudomonas aeruginosa in breath of children with cystic fibrosis. Pediatr Pulmonol, 44(2), 142-147. doi:10.1002/ppul.20963
42. Berna, A.Z., McCarthy, J.S., Wang, R.S., Saliba, K.J., Bravo, F.G., Cassells, J., … & Trowell, S.C. (2015). Analysis of Breath Specimens for Biomarkers of Plasmodium falciparum Infection. The Journal of Infectious Disease, 212, 1120-1128. doi:10.1093/infdis/jiv176
43. Bos, L.D.J., Sterk, P.J., Schultz, M.J. (2013). Volatile Metabolites of Pathogens: A Systematic Review. PLoS Pathog, 9(5), e1003311.doi:10.1371/journal.ppat.1003311
44. Buszewski, B., Kesy, M., Ligor, T., & Amann, A. (2007). Human Exhaled air analytics: biormarkers of diseases. Biomed Chromatogr,21, 553-566. doi:10.1002/bmc.835
45. Mayr, D., Margesin, R., Klingsbichel, E., Hartungen, E., Jenewein, D., Schinner, F., & Märk, T.D. (2003). Rapid Detection of Meat Spoilage by Measuring Volatile Organic Compounds by Using Proton Transfer Reaction Mass Spectrometry. Appl Environ Microbiol, 69(8), 4697-4705. doi:10.1128/AEM.69.8.4697-4705.2003
46. Wilson, A., and Baietto, M. (2009). Applications and Advanaces in Electronic-Nose Technologies. Sensors, 9(7), 5099-5148.doi:10.3390/s90705099
47. Wilson, A. (2015). Advances in Electronic- Nose Technologies for the Detection of Volatile Biomarker Metabolites in the Human Breath. Metabolites, 5, 140-163.doi:10.3390/metabo5010140
48. Spencer, T.L, Lee, A.B., & Hu, D.L. (2018). A Biomimetic Nose for Advanced Threat Detection. HDAIC Journal, 5(1), 11-15. Retrieved from https://www.hdiac.org/wp-content/uploads/2018/04/A_Biomimetic_Nose_for_Advanced_Threat_Detection_V5I1_0.pdf
49. Giannoukos, S., Agapiou, A., Taylor, S.(2018, January). Advances in chemicalsensing technologies for VOCs in breath for security/threat assessment, illicit drug detection, and human trafficking activity. Journal of Breath Research, 12(2). doi:10.1088/1752- 7163/aa95dd
50. Angle, C., Waggoner, L.P., Ferrando, A., Haney, P., & Passler, T. (2016). Canine Detection of the Volatilome: A Review of Implications for Pathogen and Disease Detection. Frontiers in Veterinary Science, 3(47). doi:10.3389/fvets.2016.00047.
51. Staymates, M.E., MacCrehan, W.A., Staymates, J.L., Kunz, R.R., Mendum, T., Ong, T.H., … & Craven B.A. (2016). Biomimetic Sniffing Improves the Detection Performance of a 3D Printed Nose of a Dog and a Commercial Trace Vapor Detetor. Scientific Reports, 6, 1-10. doi:10.1038/srep36876
52. Wilson, A.D., & Baietto, M..(2011). Advances in Electronic-Nose Technologies Developed for Biomedical Applications. Sensors, 11, 1105-1176. doi:10.3390/s110101105
53. Wilson, A.D. (2015). Advances in Electronic- Nose Technologies for the Detection of Volatile Biomarker Metabolites in Human Breath. Metabolites, 5, 140-163. doi:10.3390/metabo5010140
54. Vargas, C., Wilhelm, A.A., Williams, J., Lucas, P., Reynolds, K.A., Riley, M.R. (2009). Appl.Environ.Microbiol, 75(20), 6431-6440.doi:10.1128/AEM.02036-08
55. Priest, P.C., Duncan, A.R., Jennings, L.C., & Baker, M.G. (2011). Thermal Image Scanning for Influenza Border Screening: Results of an Airport Screening Study. PloS ONE, 6(1), e14490. doi:10.1371/journal.pone.0014490
56. St. John, R.K., King, A., Jong, D., Bodie- Collins, M., Squires, S.G., Tam, T.W.S. (2005). Border Screening for SARS. Emerging Infectious Diseases, 11(1), 6-10. Retrieved from https://wwwnc.cdc.gov/eid/article/11/1/04-0835_article
57. Bitar, D., Goubar, A., Desenclos, J.C. (2008). International travels and fever screening during epdimics: a literature review on the effectiveness and potential use of non-contact infrared thermometers. Euro Surveillance, 14(6), 1-5. doi: 10.2807/ese.14.06.19115-en
58. Michel, A.P.M., Liakat, S., Bors, K., & Gmachl, C.F. (2013). In vivo measurement of mid-infrared light scattinering from human skin.Biomedical Optics Express, 4(4), 1-11.doi: 10.1364/BOE.4.000520
59. Zarnowiec, P., Lechowicz, L., Czwerwonka, G., & Kaca, W. (2015). Fourier Transform Infrared Spectroscopy (FTIR) as a Tool for Identification and Differentiation of Pathogenic Bacteria. Current Medicinal Chemistry, 22(14), 1710-1718. doi:10.2174/0929867322666150311152800
60. Al-holy, M.A., Lin, M., Cavinato, A.G., & Rasco, B.A. (2006). The use of Fourier transform infrared spectroscopy to differentiate Escherichia coli O157:H7 from other bacteria
inoculated into apple juice. Food Microbiology, 23(2), 162-168. doi:10.1016./j.fm.2005.01.017
61. Bruyne, S.D., Speeckaert, M.M., & Delanghe, J.R. (2017). Applications of mid-infrared spectroscopy in the clinical laboratory setting. Critical Reviews in Clinical Laboratory Sciences, 55(1). doi:10.1080/10408363.2017.1414142
62. Bosch, A., Miñán, A., Vescina, C., Degrossi, J., Gatti, B., Montanaro, P., … & Yantorno, O. (2008). Fourier Transform Infrared Spectroscopy for Rapid Identification of Nonfermenting Gram-Negative Bacteria Isolated from Sputum Samples from Cystic Fibrosis Patients. Journal of Clinical Microbiology, 46(8), 2535-2546. doi:10.1128/JCM.02267-07
63. Bhunia, A.K. (2008). Chapter 1-Biosensors and Bio-Based Methods for the Separation and Detection of Foodborne Pathogens. Advances in Food and Nutrition Research, 54, 1-44. doi:10.1016/S1043-4526(07)00001-0\
64. Goodarzi, M. and Saeys, W. (2016). Selection of the most informative near infrared spectroscopy wavebands for continuous glucose monitoring in human serum. Science Direct, 146(1), 155-165. doi: 10.1016/j.talanta.2015.08.033
65. Fernandes, J. N., dos Santos, L.M.B., Chouin- Carneiro, T., Pavan, M.G., Garcia, G.A., David, M. R., … & Sikulu-Lord, M.T. (2018). Rapid, noninvasive detection of Zika virus in Aedes aegypti mosquitoes by near-infrared spectroscopy. Science Advances, 4(5), 1-6. doi: 10.1126/sciadv.aat0496
66. Tsang, M.K., Ye, W., Wang, G., Li, J., Yang, M., & Hao, J. (2016). Ultrasensitive Detection of Ebola Virus Olignonucleotide Basedon Upconversion Nanoprobe/NanoporousMembrane System. ACS Nano, 10(1), 598-605. doi: 10.1021/acsnano.5b05622


